User login
Mapping Pathology Work Associated With Precision Oncology Testing
Mapping Pathology Work Associated With Precision Oncology Testing
Comprehensive genomic profiling (CGP) is becoming progressively common and appropriate as the array of molecular targets expands. However, most hospital laboratories in the United States do not perform CGP assays in-house; instead, these tests are sent to reference laboratories. As evidenced by Inal et al, only a minority of guideline-indicated molecular testing is performed.1
The workload associated with referral testing is a barrier to increased use of such tests; streamlined processes in pathology might increase molecular test use. At 6 high-complexity US Department of Veterans Affairs (VA) medical centers (VAMCs) (Manhattan, Los Angeles, San Diego, Denver, Kansas City, and Salisbury, Maryland) ranging from 150 to 750 beds, a consult process for anatomic pathology molecular testing has increased test utilization, appropriateness of orders, standardization of reporting, and efficiency of care. This report comprehensively describes and maps the anatomic pathology molecular testing consult process at a VAMC. We present areas of inefficiency and a target state process map that incorporates best practices.
MOLECULAR TESTING CONSULT PROCESS
At the Kansas City VAMC (KCVAMC), a consult process for anatomic pathology molecular testing was introduced in 2021. Prior to this, requesting anatomic pathology molecular testing was not standardized. A variety of opportunities and methods were used for requests (eg, phone, page, Teams message, email, Computerized Patient Record System alert; or in-person during tumor board, an office meeting, or in passing). Requests were not documented in a standardized way, resulting in duplicate requests. Testing status and updates were documented outside the medical record, so requests for status updates (via various opportunities and methods) were common and redundant. Data from the year preceding consult implementation and the year following consult implementation have demonstrated increased test utilization, appropriateness of orders, standardization of reporting, and efficiency of care.
Consult Request
The precision oncology testing process starts with a health care practitioner (HCP) request on behalf of any physician or advanced practice registered nurse. It can be placed by any health care employee and directed to a designated employee in the pathology department. The request is ultimately reviewed by a pathologist (Figure 1). At KCVAMC, this request comes in the form of a consult in the electronic health record (EHR) from the ordering HCP to a pathologist. The KCVAMC pathology consult form was previously published with a discussion of the rationale for this process as opposed to a laboratory order process.2 This consult form ensures ordering HCPs supply all necessary information for the pathologist to approve the request and order the test without needing to, in most cases, contact the ordering HCP for clarification or additional information. The form asks the ordering HCP to specify which test is being requested and why. Within the Veterans Health Administration (VHA) there are local and national contracts with many laboratories with hundreds of precision oncology tests to choose from. Consulting with a pathologist is necessary to determine which test is most appropriate.
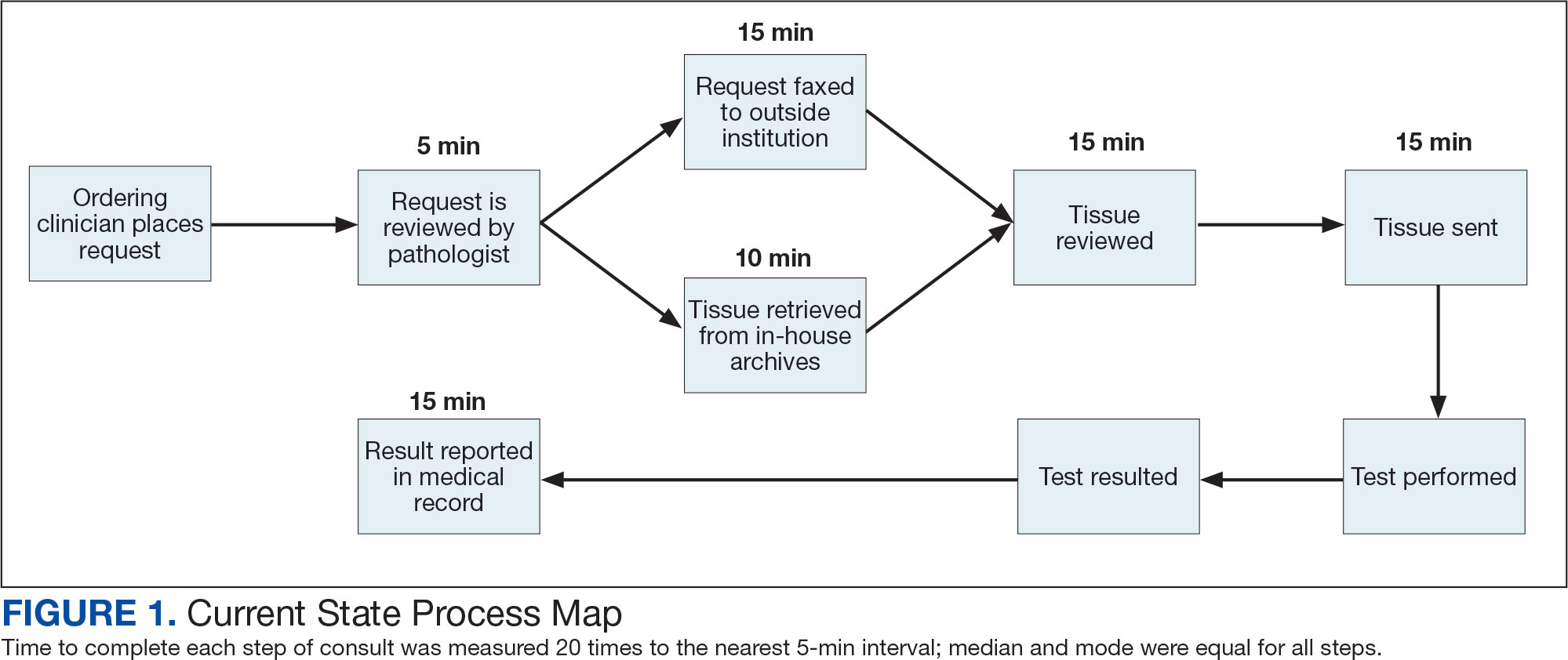
The precision oncology consult form cannot be submitted without completing all required fields. It also contains indications for the test the ordering HCP selects to minimize unintentionally inappropriate orders. The form asks which tissue the requestor expects the test to be performed on. The requestor must provide contact information for the originating institution when the tissue was collected outside the VHA. The consult form also asks whether another anatomic site is accessible and could be biopsied without unacceptable risk or impracticality, should all previously collected tissue be insufficient. For CGP requests, this allows the pathologist to determine the appropriateness of liquid biopsy without having to reach out to the ordering HCP or wait for the question to be addressed at a tumor board. When a companion diagnostic is available for a test, the ordering HCP is asked which drug will be used so that the most appropriate assay is chosen.
Consult Review
Pathology service involvement begins with pathologist review of the consult form to ensure that the correct test is indicated. Depending on the resources and preferences at a site, consults can be directed to and reviewed by the pathologist associated with the corresponding pathology specimen or to a single pathologist or group of pathologists charged with attending to consults.
The patient’s EHR is reviewed to verify that the test has not already been performed and to determine which tissue to review. Previous surgical pathology reports are examined to assess whether sufficient tissue is available for testing, which may be determined without the need for direct slide examination. Pathologists often use wording such as “rare cells” or in some cases specify that there are not enough lesional cells for ancillary testing. In biopsy reports, the percentage of tissue occupied by lesional cells or the greatest linear length of tumor cells is often documented. As for quality, pathologists may note that a specimen is largely necrotic, and gross descriptions will indicate if a specimen was compromised for molecular analysis by exposure to fixatives such as Bouin’s solution, B-5, or decalcifying agents that contain strong acids.
Tissue Retrieval
If, after such evaluation, the test is indicated and there is tissue that could be sufficient for testing, retrieval of the tissue is pursued. For in-house cases, the pathologist reviews the corresponding surgical pathology report to determine which blocks and slides to pull from the archives. In the cancer checklist, some pathologists specify the best block for subsequent ancillary studies. From the final diagnosis and gross description, the pathologist can determine which blocks are most likely to contain lesional tissue. These slides are retrieved from the archives.
For cases collected at an outside institution (other VHA facility or non-VHA facility/institution), the outside institution must be contacted to retrieve the needed slides and blocks. The phone numbers, fax numbers, email addresses, and mailing addresses for outside institutions are housed in an electronic file and are specific to the point of contact for such requests. Maintaining a record of contacts increases efficiency of the overall process; gathering contact information and successfully requesting tissue often involves multiple automated answering systems, misdirected calls, and failed attempts.
Tissue Review
After retrieving in-house tissue, the pathologist can proceed directly to slide review. For outside cases, the case must first be accessioned so that after review of the slides the pathologist can issue a report to confirm the outside diagnosis. In reviewing the slides, the pathologist looks to see that the diagnosis is correct, that there is a sufficient number of lesional cells in a section, that the lesional cells are of a sufficient concentration in a section, or subsection of the section that could be dissected, and that the cells are viable. Depending on the requested assay and the familiarity of the pathologist with that assay, the pathologist may need to look up the technical requirements of the assay and capabilities of the testing company. Assays vary in sensitivity and require differing amounts and concentrations of tumor. Some companies will dissect tissue, others will not.
If there is sufficient tissue in the material reviewed, the corresponding blocks are retrieved from in-house archives or requests are placed for outside blocks or unstained slides. If there was not enough tissue for testing, the same process is repeated to retrieve and evaluate any other specimens the patient may have. If there are no other specimens to review, this is simply communicated to the ordering HCP via the consult. If the patient is a candidate for liquid biopsy—ie, current specimens are of insufficient quality and/or quantity and a new tissue sample cannot be obtained due to unacceptable risk or impracticality—the order is placed at this time.
Tissue Transport and Testing
Unstained slides need to be cut unless blocks are sent. Slides, blocks, reports, and requisition forms are packaged for transport. An accession number is created for the precision oncology molecular laboratory test in the clinical laboratory section of the EHR system. The clinical laboratory accession number provides a way of tracking sendout testing status. The case is accessioned just prior to placement in the mail so that when an accession number appears in the EHR, the ordering HCP knows the case has been sent out. When results are received, the clinical laboratory accession is completed and a comment is added to indicate where in the EHR to find the report or, when applicable, notes that testing failed.
RESULT REPORTING
When a result becomes available, the report file is downloaded from the vendor portal. This full report is securely transmitted to the ordering HCP. The file is then scanned into the EHR. Additionally, salient findings from the report are abstracted by the pathologist for inclusion as a supplement to the anatomic pathology case. This step ensures that this information travels with the anatomic pathology report if the patient’s care is transferred elsewhere. Templates are used to ensure essential data is captured based on the type of test. The template reminds the pathologist to comment on things such as variants that may represent clonal hematopoiesis, variants that may be germline, and variants that qualify a patient for germline testing. Even with the template, the pathologist must spend significant time reviewing the chart for things such as personal cancer history, other medical history, other masses on imaging, family history, previous surgical pathology reports, and previous molecular testing.
If results are suboptimal, recommendations for repeat testing are made based on the consult response to the question of repeat biopsy feasibility and review of previous pathology reports. The final consult report is added as a consult note, the consult is completed, and the original vendor report file is associated with the consult note in the EHR.
Ancillary Testing Technician
Due to chronic KCVAMC understaffing in the clerical office, gross room, and histology, most of the consult tasks are performed by a pathologist. In an ideal scenario, the pathology staff would divide its time between a pathologist and another dedicated laboratory position, such as an ancillary testing technician (ATT). The ATT can assume responsibilities that do not require the expertise of a pathologist (Figure 2). In such a process, the only steps that would require a pathologist would be review of requests and slides and completion of the interpretive report. All other steps could be accomplished by someone who lacks certifications, laboratory experience, or postsecondary education.
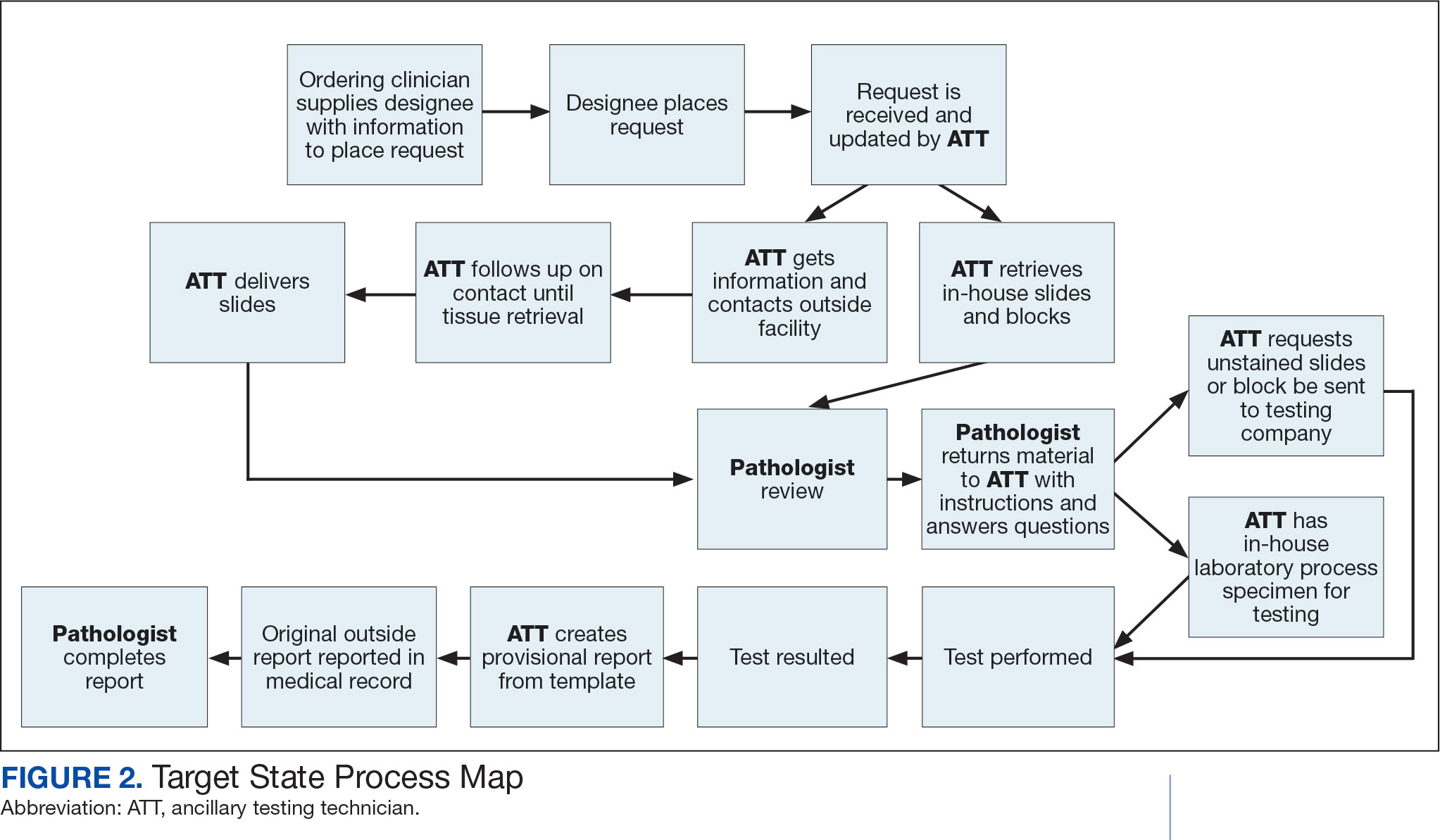
The ATT can receive the requests and retrieve slides and blocks. After slides have been reviewed by a pathologist, the pathologist can inform the ATT which slides or blocks testing will be performed on, provide any additional necessary information for completing the order, and answer any questions. For send-out tests, this allows the ATT to independently complete online portal forms and all other physical requirements prior to delivery of the slides and blocks to specimen processors in the laboratory.
ATTs can keep the ordering HCPs informed of status and be identified as the point of contact for all status inquiries. ATTs can receive results and get outside reports scanned into the EHR. Finally, ATTs can use pathologistdesigned templates to transpose information from outside reports such that a provisional report is prepared and a pathologist does not spend time duplicating information from the outside report. The pathologist can then complete the report with information requiring medical judgment that enhances care.
Optimal Pathologist Involvement
Only 3 steps in the process (request review, tissue review, and completion of an interpretive report) require a pathologist, which are necessary for optimal care and to address barriers to precision oncology.3 While the laboratory may consume only 5% of a health system budget, optimal laboratory use could prevent as much as 30% of avoidable costs.4 These estimates are widely recognized and addressed by campaigns such as Choosing Wisely, as well as programming of alerts and hard stops in EHR systems to reduce duplicate or otherwise inappropriate orders. The tests associated with precision oncology, such as CGP assays, require more nuanced consideration that is best achieved through pathology consultation. In vetting requests for such tests, the pathologist needs information that ordering HCPs do not routinely provide when ordering other tests. A consult asking for such information allows an ordering HCP to efficiently convey this information without having to call the laboratory to circumvent a hard stop.
Regardless of whether a formal electronic consult is used, pathologists must be involved in the review of requests. Creation of an original in-house report also provides an opportunity for pathologists to offer their expertise and maximize the contribution of pathology to patient care. If outside (other VHA facility or non-VHA facility/institution) reports are simply scanned into the EHR without review and issuance of an interpretive report by an in-house pathologist, then an interpretation by a pathologist with access to the patient’s complete chart is never provided. Testing companies are not provided with every patient diagnosis, so in patients with multiple neoplastic conditions, a report may seem to indicate that a detected mutation is from 1 tumor when it is actually from another. Even when all known diagnoses are considered, a variant may be detected that the medical record could reveal to indicate a new diagnosis.
Variation in reporting between companies necessitates pathologist review to standardize care. Some companies indicate which variants may represent clonal hematopoiesis, while others will simply list the pathogenic variants. An oncologist who sees a high volume of hematolymphoid neoplasia may recognize which variants may represent clonal hematopoiesis, but others may not. Reports from the same company may vary, and their interpretation often requires a pathologist's expertise. For example, even if a sample meets the technical requirements for analysis, the report may indicate that the quality or quantity of DNA has reduced the sensitivity for genomic alteration detection. A pathologist would know how to use this information in deciding how to proceed. In a situation where quantity was the issue, the pathologist may know there is additional tissue that could be sent for testing. If quality is the issue, the pathologist may know that additional blocks from the same case likely have the same quality of DNA and would also be unsuitable for testing.
Pathologist input is necessary for precision oncology testing. Some tasks that would ideally be completed by a molecular pathologist (eg, creation of reports to indicate which variants may represent clonal hematopoiesis of indeterminate potential) may be sufficiently completed by a pathologist without fellowship training in molecular pathology.
There are about 15,000 full-time pathologists in the US.4 In the 20 years since molecular genetic pathology was formally recognized as a specialty, there have been < 500 pathologists who have pursued fellowship training in this specialty.5 With the inundation of molecular variants uncovered by routine next-generation sequencing (NGS), there are too few fellowship-trained molecular pathologists to provide all such aforementioned input; it is incumbent on surgical pathologists in general to take on such responsibilities.
Consult Implementation Data
These results support the feasibility and effectiveness of the consult process. Prior to consult implementation, many requests were not compliant with VHA National Precision Oncology Program (NPOP) testing guidelines. Since enactment of the consult, > 90% of requests have been in compliance. In the year preceding the consult (January 2020 to December 2021), 55 of 211 (26.1%) metastatic lung and prostate cancers samples eligible for NGS were tested and 126 (59.7%) NGS vendor reports were scanned into the EHR. The mean time from metastasis to NGS result was 151 days. In the year following enactment of the consult (January 2021 to December 2022), 168 of 224 (75.0%) of metastatic lung and prostate cancers eligible for NGS were tested and all 224 NGS vendor reports were scanned into the EHR. The mean time from metastasis to NGS result was 83 days. These data indicate that the practices recommended increase test use, appropriateness of orders, standardization of reporting, and efficiency of care.
CONCLUSIONS
Processing precision oncology testing requires substantial work for pathology departments. Laboratory workforce shortages and ever-expanding indications necessitate additional study of pathology processes to manage increasing workload and maintain the highest quality of cancer care through maximal efficiency and the development of appropriate staffing models. The use of a consult for anatomic pathology molecular testing is one process that can increase test use, appropriateness of orders, standardization of reporting, and efficiency of care. This report provides a comprehensive description and mapping of the process, highlights best practices, identifies inefficiencies, and provides a description and mapping of a target state.
- Inal C, Yilmaz E, Cheng H, et al. Effect of reflex testing by pathologists on molecular testing rates in lung cancer patients: experience from a community-based academic center. J Clin Oncol. 2014;32(15 suppl):8098. doi:10.1200/jco.2014.32.15_suppl.8098
- Mettman D, Goodman M, Modzelewski J, et al. Streamlining institutional pathway processes: the development and implementation of a pathology molecular consult to facilitate convenient and efficient ordering, fulfillment, and reporting for tissue molecular tests. J Clin Pathw.Ersek JL, Black LJ, Thompson MA, Kim ES. Implementing precision medicine programs and clinical trials in the community-based oncology practice: barriers and best practices. Am Soc Clin Oncol Educ Book. 2018;38:188- 196. doi:10.1200/EDBK_200633 2022;8(1):28-33.
- Ersek JL, Black LJ, Thompson MA, Kim ES. Implementing precision medicine programs and clinical trials in the community-based oncology practice: barriers and best practices. Am Soc Clin Oncol Educ Book. 2018;38:188- 196. doi:10.1200/EDBK_200633
- Robboy SJ, Gupta S, Crawford JM, et al. The pathologist workforce in the United States: II. An interactive modeling tool for analyzing future qualitative and quantitative staffing demands for services. Arch Pathol Lab Med. 2015;139(11):1413-1430. doi:10.5858/arpa.2014-0559-OA doi:10.25270/jcp.2022.02.1
- Robboy SJ, Gross D, Park JY, et al. Reevaluation of the US pathologist workforce size. JAMA Netw Open. 2020;3(7): e2010648. doi:10.1001/jamanetworkopen.2020.10648
Comprehensive genomic profiling (CGP) is becoming progressively common and appropriate as the array of molecular targets expands. However, most hospital laboratories in the United States do not perform CGP assays in-house; instead, these tests are sent to reference laboratories. As evidenced by Inal et al, only a minority of guideline-indicated molecular testing is performed.1
The workload associated with referral testing is a barrier to increased use of such tests; streamlined processes in pathology might increase molecular test use. At 6 high-complexity US Department of Veterans Affairs (VA) medical centers (VAMCs) (Manhattan, Los Angeles, San Diego, Denver, Kansas City, and Salisbury, Maryland) ranging from 150 to 750 beds, a consult process for anatomic pathology molecular testing has increased test utilization, appropriateness of orders, standardization of reporting, and efficiency of care. This report comprehensively describes and maps the anatomic pathology molecular testing consult process at a VAMC. We present areas of inefficiency and a target state process map that incorporates best practices.
MOLECULAR TESTING CONSULT PROCESS
At the Kansas City VAMC (KCVAMC), a consult process for anatomic pathology molecular testing was introduced in 2021. Prior to this, requesting anatomic pathology molecular testing was not standardized. A variety of opportunities and methods were used for requests (eg, phone, page, Teams message, email, Computerized Patient Record System alert; or in-person during tumor board, an office meeting, or in passing). Requests were not documented in a standardized way, resulting in duplicate requests. Testing status and updates were documented outside the medical record, so requests for status updates (via various opportunities and methods) were common and redundant. Data from the year preceding consult implementation and the year following consult implementation have demonstrated increased test utilization, appropriateness of orders, standardization of reporting, and efficiency of care.
Consult Request
The precision oncology testing process starts with a health care practitioner (HCP) request on behalf of any physician or advanced practice registered nurse. It can be placed by any health care employee and directed to a designated employee in the pathology department. The request is ultimately reviewed by a pathologist (Figure 1). At KCVAMC, this request comes in the form of a consult in the electronic health record (EHR) from the ordering HCP to a pathologist. The KCVAMC pathology consult form was previously published with a discussion of the rationale for this process as opposed to a laboratory order process.2 This consult form ensures ordering HCPs supply all necessary information for the pathologist to approve the request and order the test without needing to, in most cases, contact the ordering HCP for clarification or additional information. The form asks the ordering HCP to specify which test is being requested and why. Within the Veterans Health Administration (VHA) there are local and national contracts with many laboratories with hundreds of precision oncology tests to choose from. Consulting with a pathologist is necessary to determine which test is most appropriate.

The precision oncology consult form cannot be submitted without completing all required fields. It also contains indications for the test the ordering HCP selects to minimize unintentionally inappropriate orders. The form asks which tissue the requestor expects the test to be performed on. The requestor must provide contact information for the originating institution when the tissue was collected outside the VHA. The consult form also asks whether another anatomic site is accessible and could be biopsied without unacceptable risk or impracticality, should all previously collected tissue be insufficient. For CGP requests, this allows the pathologist to determine the appropriateness of liquid biopsy without having to reach out to the ordering HCP or wait for the question to be addressed at a tumor board. When a companion diagnostic is available for a test, the ordering HCP is asked which drug will be used so that the most appropriate assay is chosen.
Consult Review
Pathology service involvement begins with pathologist review of the consult form to ensure that the correct test is indicated. Depending on the resources and preferences at a site, consults can be directed to and reviewed by the pathologist associated with the corresponding pathology specimen or to a single pathologist or group of pathologists charged with attending to consults.
The patient’s EHR is reviewed to verify that the test has not already been performed and to determine which tissue to review. Previous surgical pathology reports are examined to assess whether sufficient tissue is available for testing, which may be determined without the need for direct slide examination. Pathologists often use wording such as “rare cells” or in some cases specify that there are not enough lesional cells for ancillary testing. In biopsy reports, the percentage of tissue occupied by lesional cells or the greatest linear length of tumor cells is often documented. As for quality, pathologists may note that a specimen is largely necrotic, and gross descriptions will indicate if a specimen was compromised for molecular analysis by exposure to fixatives such as Bouin’s solution, B-5, or decalcifying agents that contain strong acids.
Tissue Retrieval
If, after such evaluation, the test is indicated and there is tissue that could be sufficient for testing, retrieval of the tissue is pursued. For in-house cases, the pathologist reviews the corresponding surgical pathology report to determine which blocks and slides to pull from the archives. In the cancer checklist, some pathologists specify the best block for subsequent ancillary studies. From the final diagnosis and gross description, the pathologist can determine which blocks are most likely to contain lesional tissue. These slides are retrieved from the archives.
For cases collected at an outside institution (other VHA facility or non-VHA facility/institution), the outside institution must be contacted to retrieve the needed slides and blocks. The phone numbers, fax numbers, email addresses, and mailing addresses for outside institutions are housed in an electronic file and are specific to the point of contact for such requests. Maintaining a record of contacts increases efficiency of the overall process; gathering contact information and successfully requesting tissue often involves multiple automated answering systems, misdirected calls, and failed attempts.
Tissue Review
After retrieving in-house tissue, the pathologist can proceed directly to slide review. For outside cases, the case must first be accessioned so that after review of the slides the pathologist can issue a report to confirm the outside diagnosis. In reviewing the slides, the pathologist looks to see that the diagnosis is correct, that there is a sufficient number of lesional cells in a section, that the lesional cells are of a sufficient concentration in a section, or subsection of the section that could be dissected, and that the cells are viable. Depending on the requested assay and the familiarity of the pathologist with that assay, the pathologist may need to look up the technical requirements of the assay and capabilities of the testing company. Assays vary in sensitivity and require differing amounts and concentrations of tumor. Some companies will dissect tissue, others will not.
If there is sufficient tissue in the material reviewed, the corresponding blocks are retrieved from in-house archives or requests are placed for outside blocks or unstained slides. If there was not enough tissue for testing, the same process is repeated to retrieve and evaluate any other specimens the patient may have. If there are no other specimens to review, this is simply communicated to the ordering HCP via the consult. If the patient is a candidate for liquid biopsy—ie, current specimens are of insufficient quality and/or quantity and a new tissue sample cannot be obtained due to unacceptable risk or impracticality—the order is placed at this time.
Tissue Transport and Testing
Unstained slides need to be cut unless blocks are sent. Slides, blocks, reports, and requisition forms are packaged for transport. An accession number is created for the precision oncology molecular laboratory test in the clinical laboratory section of the EHR system. The clinical laboratory accession number provides a way of tracking sendout testing status. The case is accessioned just prior to placement in the mail so that when an accession number appears in the EHR, the ordering HCP knows the case has been sent out. When results are received, the clinical laboratory accession is completed and a comment is added to indicate where in the EHR to find the report or, when applicable, notes that testing failed.
RESULT REPORTING
When a result becomes available, the report file is downloaded from the vendor portal. This full report is securely transmitted to the ordering HCP. The file is then scanned into the EHR. Additionally, salient findings from the report are abstracted by the pathologist for inclusion as a supplement to the anatomic pathology case. This step ensures that this information travels with the anatomic pathology report if the patient’s care is transferred elsewhere. Templates are used to ensure essential data is captured based on the type of test. The template reminds the pathologist to comment on things such as variants that may represent clonal hematopoiesis, variants that may be germline, and variants that qualify a patient for germline testing. Even with the template, the pathologist must spend significant time reviewing the chart for things such as personal cancer history, other medical history, other masses on imaging, family history, previous surgical pathology reports, and previous molecular testing.
If results are suboptimal, recommendations for repeat testing are made based on the consult response to the question of repeat biopsy feasibility and review of previous pathology reports. The final consult report is added as a consult note, the consult is completed, and the original vendor report file is associated with the consult note in the EHR.
Ancillary Testing Technician
Due to chronic KCVAMC understaffing in the clerical office, gross room, and histology, most of the consult tasks are performed by a pathologist. In an ideal scenario, the pathology staff would divide its time between a pathologist and another dedicated laboratory position, such as an ancillary testing technician (ATT). The ATT can assume responsibilities that do not require the expertise of a pathologist (Figure 2). In such a process, the only steps that would require a pathologist would be review of requests and slides and completion of the interpretive report. All other steps could be accomplished by someone who lacks certifications, laboratory experience, or postsecondary education.

The ATT can receive the requests and retrieve slides and blocks. After slides have been reviewed by a pathologist, the pathologist can inform the ATT which slides or blocks testing will be performed on, provide any additional necessary information for completing the order, and answer any questions. For send-out tests, this allows the ATT to independently complete online portal forms and all other physical requirements prior to delivery of the slides and blocks to specimen processors in the laboratory.
ATTs can keep the ordering HCPs informed of status and be identified as the point of contact for all status inquiries. ATTs can receive results and get outside reports scanned into the EHR. Finally, ATTs can use pathologistdesigned templates to transpose information from outside reports such that a provisional report is prepared and a pathologist does not spend time duplicating information from the outside report. The pathologist can then complete the report with information requiring medical judgment that enhances care.
Optimal Pathologist Involvement
Only 3 steps in the process (request review, tissue review, and completion of an interpretive report) require a pathologist, which are necessary for optimal care and to address barriers to precision oncology.3 While the laboratory may consume only 5% of a health system budget, optimal laboratory use could prevent as much as 30% of avoidable costs.4 These estimates are widely recognized and addressed by campaigns such as Choosing Wisely, as well as programming of alerts and hard stops in EHR systems to reduce duplicate or otherwise inappropriate orders. The tests associated with precision oncology, such as CGP assays, require more nuanced consideration that is best achieved through pathology consultation. In vetting requests for such tests, the pathologist needs information that ordering HCPs do not routinely provide when ordering other tests. A consult asking for such information allows an ordering HCP to efficiently convey this information without having to call the laboratory to circumvent a hard stop.
Regardless of whether a formal electronic consult is used, pathologists must be involved in the review of requests. Creation of an original in-house report also provides an opportunity for pathologists to offer their expertise and maximize the contribution of pathology to patient care. If outside (other VHA facility or non-VHA facility/institution) reports are simply scanned into the EHR without review and issuance of an interpretive report by an in-house pathologist, then an interpretation by a pathologist with access to the patient’s complete chart is never provided. Testing companies are not provided with every patient diagnosis, so in patients with multiple neoplastic conditions, a report may seem to indicate that a detected mutation is from 1 tumor when it is actually from another. Even when all known diagnoses are considered, a variant may be detected that the medical record could reveal to indicate a new diagnosis.
Variation in reporting between companies necessitates pathologist review to standardize care. Some companies indicate which variants may represent clonal hematopoiesis, while others will simply list the pathogenic variants. An oncologist who sees a high volume of hematolymphoid neoplasia may recognize which variants may represent clonal hematopoiesis, but others may not. Reports from the same company may vary, and their interpretation often requires a pathologist's expertise. For example, even if a sample meets the technical requirements for analysis, the report may indicate that the quality or quantity of DNA has reduced the sensitivity for genomic alteration detection. A pathologist would know how to use this information in deciding how to proceed. In a situation where quantity was the issue, the pathologist may know there is additional tissue that could be sent for testing. If quality is the issue, the pathologist may know that additional blocks from the same case likely have the same quality of DNA and would also be unsuitable for testing.
Pathologist input is necessary for precision oncology testing. Some tasks that would ideally be completed by a molecular pathologist (eg, creation of reports to indicate which variants may represent clonal hematopoiesis of indeterminate potential) may be sufficiently completed by a pathologist without fellowship training in molecular pathology.
There are about 15,000 full-time pathologists in the US.4 In the 20 years since molecular genetic pathology was formally recognized as a specialty, there have been < 500 pathologists who have pursued fellowship training in this specialty.5 With the inundation of molecular variants uncovered by routine next-generation sequencing (NGS), there are too few fellowship-trained molecular pathologists to provide all such aforementioned input; it is incumbent on surgical pathologists in general to take on such responsibilities.
Consult Implementation Data
These results support the feasibility and effectiveness of the consult process. Prior to consult implementation, many requests were not compliant with VHA National Precision Oncology Program (NPOP) testing guidelines. Since enactment of the consult, > 90% of requests have been in compliance. In the year preceding the consult (January 2020 to December 2021), 55 of 211 (26.1%) metastatic lung and prostate cancers samples eligible for NGS were tested and 126 (59.7%) NGS vendor reports were scanned into the EHR. The mean time from metastasis to NGS result was 151 days. In the year following enactment of the consult (January 2021 to December 2022), 168 of 224 (75.0%) of metastatic lung and prostate cancers eligible for NGS were tested and all 224 NGS vendor reports were scanned into the EHR. The mean time from metastasis to NGS result was 83 days. These data indicate that the practices recommended increase test use, appropriateness of orders, standardization of reporting, and efficiency of care.
CONCLUSIONS
Processing precision oncology testing requires substantial work for pathology departments. Laboratory workforce shortages and ever-expanding indications necessitate additional study of pathology processes to manage increasing workload and maintain the highest quality of cancer care through maximal efficiency and the development of appropriate staffing models. The use of a consult for anatomic pathology molecular testing is one process that can increase test use, appropriateness of orders, standardization of reporting, and efficiency of care. This report provides a comprehensive description and mapping of the process, highlights best practices, identifies inefficiencies, and provides a description and mapping of a target state.
Comprehensive genomic profiling (CGP) is becoming progressively common and appropriate as the array of molecular targets expands. However, most hospital laboratories in the United States do not perform CGP assays in-house; instead, these tests are sent to reference laboratories. As evidenced by Inal et al, only a minority of guideline-indicated molecular testing is performed.1
The workload associated with referral testing is a barrier to increased use of such tests; streamlined processes in pathology might increase molecular test use. At 6 high-complexity US Department of Veterans Affairs (VA) medical centers (VAMCs) (Manhattan, Los Angeles, San Diego, Denver, Kansas City, and Salisbury, Maryland) ranging from 150 to 750 beds, a consult process for anatomic pathology molecular testing has increased test utilization, appropriateness of orders, standardization of reporting, and efficiency of care. This report comprehensively describes and maps the anatomic pathology molecular testing consult process at a VAMC. We present areas of inefficiency and a target state process map that incorporates best practices.
MOLECULAR TESTING CONSULT PROCESS
At the Kansas City VAMC (KCVAMC), a consult process for anatomic pathology molecular testing was introduced in 2021. Prior to this, requesting anatomic pathology molecular testing was not standardized. A variety of opportunities and methods were used for requests (eg, phone, page, Teams message, email, Computerized Patient Record System alert; or in-person during tumor board, an office meeting, or in passing). Requests were not documented in a standardized way, resulting in duplicate requests. Testing status and updates were documented outside the medical record, so requests for status updates (via various opportunities and methods) were common and redundant. Data from the year preceding consult implementation and the year following consult implementation have demonstrated increased test utilization, appropriateness of orders, standardization of reporting, and efficiency of care.
Consult Request
The precision oncology testing process starts with a health care practitioner (HCP) request on behalf of any physician or advanced practice registered nurse. It can be placed by any health care employee and directed to a designated employee in the pathology department. The request is ultimately reviewed by a pathologist (Figure 1). At KCVAMC, this request comes in the form of a consult in the electronic health record (EHR) from the ordering HCP to a pathologist. The KCVAMC pathology consult form was previously published with a discussion of the rationale for this process as opposed to a laboratory order process.2 This consult form ensures ordering HCPs supply all necessary information for the pathologist to approve the request and order the test without needing to, in most cases, contact the ordering HCP for clarification or additional information. The form asks the ordering HCP to specify which test is being requested and why. Within the Veterans Health Administration (VHA) there are local and national contracts with many laboratories with hundreds of precision oncology tests to choose from. Consulting with a pathologist is necessary to determine which test is most appropriate.

The precision oncology consult form cannot be submitted without completing all required fields. It also contains indications for the test the ordering HCP selects to minimize unintentionally inappropriate orders. The form asks which tissue the requestor expects the test to be performed on. The requestor must provide contact information for the originating institution when the tissue was collected outside the VHA. The consult form also asks whether another anatomic site is accessible and could be biopsied without unacceptable risk or impracticality, should all previously collected tissue be insufficient. For CGP requests, this allows the pathologist to determine the appropriateness of liquid biopsy without having to reach out to the ordering HCP or wait for the question to be addressed at a tumor board. When a companion diagnostic is available for a test, the ordering HCP is asked which drug will be used so that the most appropriate assay is chosen.
Consult Review
Pathology service involvement begins with pathologist review of the consult form to ensure that the correct test is indicated. Depending on the resources and preferences at a site, consults can be directed to and reviewed by the pathologist associated with the corresponding pathology specimen or to a single pathologist or group of pathologists charged with attending to consults.
The patient’s EHR is reviewed to verify that the test has not already been performed and to determine which tissue to review. Previous surgical pathology reports are examined to assess whether sufficient tissue is available for testing, which may be determined without the need for direct slide examination. Pathologists often use wording such as “rare cells” or in some cases specify that there are not enough lesional cells for ancillary testing. In biopsy reports, the percentage of tissue occupied by lesional cells or the greatest linear length of tumor cells is often documented. As for quality, pathologists may note that a specimen is largely necrotic, and gross descriptions will indicate if a specimen was compromised for molecular analysis by exposure to fixatives such as Bouin’s solution, B-5, or decalcifying agents that contain strong acids.
Tissue Retrieval
If, after such evaluation, the test is indicated and there is tissue that could be sufficient for testing, retrieval of the tissue is pursued. For in-house cases, the pathologist reviews the corresponding surgical pathology report to determine which blocks and slides to pull from the archives. In the cancer checklist, some pathologists specify the best block for subsequent ancillary studies. From the final diagnosis and gross description, the pathologist can determine which blocks are most likely to contain lesional tissue. These slides are retrieved from the archives.
For cases collected at an outside institution (other VHA facility or non-VHA facility/institution), the outside institution must be contacted to retrieve the needed slides and blocks. The phone numbers, fax numbers, email addresses, and mailing addresses for outside institutions are housed in an electronic file and are specific to the point of contact for such requests. Maintaining a record of contacts increases efficiency of the overall process; gathering contact information and successfully requesting tissue often involves multiple automated answering systems, misdirected calls, and failed attempts.
Tissue Review
After retrieving in-house tissue, the pathologist can proceed directly to slide review. For outside cases, the case must first be accessioned so that after review of the slides the pathologist can issue a report to confirm the outside diagnosis. In reviewing the slides, the pathologist looks to see that the diagnosis is correct, that there is a sufficient number of lesional cells in a section, that the lesional cells are of a sufficient concentration in a section, or subsection of the section that could be dissected, and that the cells are viable. Depending on the requested assay and the familiarity of the pathologist with that assay, the pathologist may need to look up the technical requirements of the assay and capabilities of the testing company. Assays vary in sensitivity and require differing amounts and concentrations of tumor. Some companies will dissect tissue, others will not.
If there is sufficient tissue in the material reviewed, the corresponding blocks are retrieved from in-house archives or requests are placed for outside blocks or unstained slides. If there was not enough tissue for testing, the same process is repeated to retrieve and evaluate any other specimens the patient may have. If there are no other specimens to review, this is simply communicated to the ordering HCP via the consult. If the patient is a candidate for liquid biopsy—ie, current specimens are of insufficient quality and/or quantity and a new tissue sample cannot be obtained due to unacceptable risk or impracticality—the order is placed at this time.
Tissue Transport and Testing
Unstained slides need to be cut unless blocks are sent. Slides, blocks, reports, and requisition forms are packaged for transport. An accession number is created for the precision oncology molecular laboratory test in the clinical laboratory section of the EHR system. The clinical laboratory accession number provides a way of tracking sendout testing status. The case is accessioned just prior to placement in the mail so that when an accession number appears in the EHR, the ordering HCP knows the case has been sent out. When results are received, the clinical laboratory accession is completed and a comment is added to indicate where in the EHR to find the report or, when applicable, notes that testing failed.
RESULT REPORTING
When a result becomes available, the report file is downloaded from the vendor portal. This full report is securely transmitted to the ordering HCP. The file is then scanned into the EHR. Additionally, salient findings from the report are abstracted by the pathologist for inclusion as a supplement to the anatomic pathology case. This step ensures that this information travels with the anatomic pathology report if the patient’s care is transferred elsewhere. Templates are used to ensure essential data is captured based on the type of test. The template reminds the pathologist to comment on things such as variants that may represent clonal hematopoiesis, variants that may be germline, and variants that qualify a patient for germline testing. Even with the template, the pathologist must spend significant time reviewing the chart for things such as personal cancer history, other medical history, other masses on imaging, family history, previous surgical pathology reports, and previous molecular testing.
If results are suboptimal, recommendations for repeat testing are made based on the consult response to the question of repeat biopsy feasibility and review of previous pathology reports. The final consult report is added as a consult note, the consult is completed, and the original vendor report file is associated with the consult note in the EHR.
Ancillary Testing Technician
Due to chronic KCVAMC understaffing in the clerical office, gross room, and histology, most of the consult tasks are performed by a pathologist. In an ideal scenario, the pathology staff would divide its time between a pathologist and another dedicated laboratory position, such as an ancillary testing technician (ATT). The ATT can assume responsibilities that do not require the expertise of a pathologist (Figure 2). In such a process, the only steps that would require a pathologist would be review of requests and slides and completion of the interpretive report. All other steps could be accomplished by someone who lacks certifications, laboratory experience, or postsecondary education.

The ATT can receive the requests and retrieve slides and blocks. After slides have been reviewed by a pathologist, the pathologist can inform the ATT which slides or blocks testing will be performed on, provide any additional necessary information for completing the order, and answer any questions. For send-out tests, this allows the ATT to independently complete online portal forms and all other physical requirements prior to delivery of the slides and blocks to specimen processors in the laboratory.
ATTs can keep the ordering HCPs informed of status and be identified as the point of contact for all status inquiries. ATTs can receive results and get outside reports scanned into the EHR. Finally, ATTs can use pathologistdesigned templates to transpose information from outside reports such that a provisional report is prepared and a pathologist does not spend time duplicating information from the outside report. The pathologist can then complete the report with information requiring medical judgment that enhances care.
Optimal Pathologist Involvement
Only 3 steps in the process (request review, tissue review, and completion of an interpretive report) require a pathologist, which are necessary for optimal care and to address barriers to precision oncology.3 While the laboratory may consume only 5% of a health system budget, optimal laboratory use could prevent as much as 30% of avoidable costs.4 These estimates are widely recognized and addressed by campaigns such as Choosing Wisely, as well as programming of alerts and hard stops in EHR systems to reduce duplicate or otherwise inappropriate orders. The tests associated with precision oncology, such as CGP assays, require more nuanced consideration that is best achieved through pathology consultation. In vetting requests for such tests, the pathologist needs information that ordering HCPs do not routinely provide when ordering other tests. A consult asking for such information allows an ordering HCP to efficiently convey this information without having to call the laboratory to circumvent a hard stop.
Regardless of whether a formal electronic consult is used, pathologists must be involved in the review of requests. Creation of an original in-house report also provides an opportunity for pathologists to offer their expertise and maximize the contribution of pathology to patient care. If outside (other VHA facility or non-VHA facility/institution) reports are simply scanned into the EHR without review and issuance of an interpretive report by an in-house pathologist, then an interpretation by a pathologist with access to the patient’s complete chart is never provided. Testing companies are not provided with every patient diagnosis, so in patients with multiple neoplastic conditions, a report may seem to indicate that a detected mutation is from 1 tumor when it is actually from another. Even when all known diagnoses are considered, a variant may be detected that the medical record could reveal to indicate a new diagnosis.
Variation in reporting between companies necessitates pathologist review to standardize care. Some companies indicate which variants may represent clonal hematopoiesis, while others will simply list the pathogenic variants. An oncologist who sees a high volume of hematolymphoid neoplasia may recognize which variants may represent clonal hematopoiesis, but others may not. Reports from the same company may vary, and their interpretation often requires a pathologist's expertise. For example, even if a sample meets the technical requirements for analysis, the report may indicate that the quality or quantity of DNA has reduced the sensitivity for genomic alteration detection. A pathologist would know how to use this information in deciding how to proceed. In a situation where quantity was the issue, the pathologist may know there is additional tissue that could be sent for testing. If quality is the issue, the pathologist may know that additional blocks from the same case likely have the same quality of DNA and would also be unsuitable for testing.
Pathologist input is necessary for precision oncology testing. Some tasks that would ideally be completed by a molecular pathologist (eg, creation of reports to indicate which variants may represent clonal hematopoiesis of indeterminate potential) may be sufficiently completed by a pathologist without fellowship training in molecular pathology.
There are about 15,000 full-time pathologists in the US.4 In the 20 years since molecular genetic pathology was formally recognized as a specialty, there have been < 500 pathologists who have pursued fellowship training in this specialty.5 With the inundation of molecular variants uncovered by routine next-generation sequencing (NGS), there are too few fellowship-trained molecular pathologists to provide all such aforementioned input; it is incumbent on surgical pathologists in general to take on such responsibilities.
Consult Implementation Data
These results support the feasibility and effectiveness of the consult process. Prior to consult implementation, many requests were not compliant with VHA National Precision Oncology Program (NPOP) testing guidelines. Since enactment of the consult, > 90% of requests have been in compliance. In the year preceding the consult (January 2020 to December 2021), 55 of 211 (26.1%) metastatic lung and prostate cancers samples eligible for NGS were tested and 126 (59.7%) NGS vendor reports were scanned into the EHR. The mean time from metastasis to NGS result was 151 days. In the year following enactment of the consult (January 2021 to December 2022), 168 of 224 (75.0%) of metastatic lung and prostate cancers eligible for NGS were tested and all 224 NGS vendor reports were scanned into the EHR. The mean time from metastasis to NGS result was 83 days. These data indicate that the practices recommended increase test use, appropriateness of orders, standardization of reporting, and efficiency of care.
CONCLUSIONS
Processing precision oncology testing requires substantial work for pathology departments. Laboratory workforce shortages and ever-expanding indications necessitate additional study of pathology processes to manage increasing workload and maintain the highest quality of cancer care through maximal efficiency and the development of appropriate staffing models. The use of a consult for anatomic pathology molecular testing is one process that can increase test use, appropriateness of orders, standardization of reporting, and efficiency of care. This report provides a comprehensive description and mapping of the process, highlights best practices, identifies inefficiencies, and provides a description and mapping of a target state.
- Inal C, Yilmaz E, Cheng H, et al. Effect of reflex testing by pathologists on molecular testing rates in lung cancer patients: experience from a community-based academic center. J Clin Oncol. 2014;32(15 suppl):8098. doi:10.1200/jco.2014.32.15_suppl.8098
- Mettman D, Goodman M, Modzelewski J, et al. Streamlining institutional pathway processes: the development and implementation of a pathology molecular consult to facilitate convenient and efficient ordering, fulfillment, and reporting for tissue molecular tests. J Clin Pathw.Ersek JL, Black LJ, Thompson MA, Kim ES. Implementing precision medicine programs and clinical trials in the community-based oncology practice: barriers and best practices. Am Soc Clin Oncol Educ Book. 2018;38:188- 196. doi:10.1200/EDBK_200633 2022;8(1):28-33.
- Ersek JL, Black LJ, Thompson MA, Kim ES. Implementing precision medicine programs and clinical trials in the community-based oncology practice: barriers and best practices. Am Soc Clin Oncol Educ Book. 2018;38:188- 196. doi:10.1200/EDBK_200633
- Robboy SJ, Gupta S, Crawford JM, et al. The pathologist workforce in the United States: II. An interactive modeling tool for analyzing future qualitative and quantitative staffing demands for services. Arch Pathol Lab Med. 2015;139(11):1413-1430. doi:10.5858/arpa.2014-0559-OA doi:10.25270/jcp.2022.02.1
- Robboy SJ, Gross D, Park JY, et al. Reevaluation of the US pathologist workforce size. JAMA Netw Open. 2020;3(7): e2010648. doi:10.1001/jamanetworkopen.2020.10648
- Inal C, Yilmaz E, Cheng H, et al. Effect of reflex testing by pathologists on molecular testing rates in lung cancer patients: experience from a community-based academic center. J Clin Oncol. 2014;32(15 suppl):8098. doi:10.1200/jco.2014.32.15_suppl.8098
- Mettman D, Goodman M, Modzelewski J, et al. Streamlining institutional pathway processes: the development and implementation of a pathology molecular consult to facilitate convenient and efficient ordering, fulfillment, and reporting for tissue molecular tests. J Clin Pathw.Ersek JL, Black LJ, Thompson MA, Kim ES. Implementing precision medicine programs and clinical trials in the community-based oncology practice: barriers and best practices. Am Soc Clin Oncol Educ Book. 2018;38:188- 196. doi:10.1200/EDBK_200633 2022;8(1):28-33.
- Ersek JL, Black LJ, Thompson MA, Kim ES. Implementing precision medicine programs and clinical trials in the community-based oncology practice: barriers and best practices. Am Soc Clin Oncol Educ Book. 2018;38:188- 196. doi:10.1200/EDBK_200633
- Robboy SJ, Gupta S, Crawford JM, et al. The pathologist workforce in the United States: II. An interactive modeling tool for analyzing future qualitative and quantitative staffing demands for services. Arch Pathol Lab Med. 2015;139(11):1413-1430. doi:10.5858/arpa.2014-0559-OA doi:10.25270/jcp.2022.02.1
- Robboy SJ, Gross D, Park JY, et al. Reevaluation of the US pathologist workforce size. JAMA Netw Open. 2020;3(7): e2010648. doi:10.1001/jamanetworkopen.2020.10648
Mapping Pathology Work Associated With Precision Oncology Testing
Mapping Pathology Work Associated With Precision Oncology Testing
Nationwide Genomic Surveillance and Response to COVID-19: The VA SeqFORCE and SeqCURE Consortiums
The COVID-19 virus and its associated pandemic have highlighted the urgent need for a national infrastructure to rapidly identify and respond to emerging pathogens. The importance of understanding viral population dynamics through genetic sequencing has become apparent over time, particularly as the vaccine responses, clinical implications, and therapeutic effectiveness of treatments have varied substantially with COVID-19 variants.1,2
As the largest integrated health care system in the US, the US Department of Veterans Affairs (VA) is uniquely situated to help with pandemic detection and response. This article highlights 2 VA programs dedicated to COVID-19 sequencing at the forefront of pandemic response and research: VA Sequencing for Research Clinical and Epidemiology (SeqFORCE) and VA Sequencing Collaborations United for Research and Epidemiology (SeqCURE) (Table).
VA Seq FORCE
VA SeqFORCE was established March 2021 to facilitate clinical surveillance of COVID-19 variants in the US veteran population and in VA employees. VA SeqFORCE consists of 9 Clinical Laboratory Improvement Amendment (CLIA)–certified laboratories in VA medical centers, including the VA Public Health Reference Laboratory in Palo Alto, California, and 8 Veterans Health Administration (VHA) clinical laboratories (Los Angeles, California; Boise, Idaho; Iowa City, Iowa; Bronx, New York; West Haven, Connecticut; Indianapolis, Indiana; Denver, Colorado; and Orlando, Florida).3 Specimen standards (eg, real-time polymerase chain reaction [RT-PCR] cycle threshold [Ct] ≤ 30, minimum volume, etc) and clinical criteria (eg, COVID-19–related deaths, COVID-19 vaccine escape, etc) for submitting samples to VA SeqFORCE laboratories were established, and logistics for sample sequencing was centralized, including providing centralized instructions for sample preparation and to which VA SeqFORCE laboratory samples should be sent.
These laboratories sequenced samples from patients and employees with COVID-19 to understand patterns of variant evolution, vaccine, antiviral and monoclonal antibody response, health care–associated outbreaks, and COVID-19 transmission. As clinically relevant findings, such as monoclonal antibody treatment failure, emerged with novel viral variants, VA SeqFORCE was well positioned to rapidly detect the emergent variants and inform better clinical care of patients with COVID-19. Other clinical indications identified for sequencing within VA SeqFORCE included outbreak investigation, re-infection with COVID-19 > 90 days but < 6 months after a prior infection, extended hospitalization of > 21 days, death due to COVID-19, infection with a history of recent nondomestic travel, rebound of symptoms after improvement on oral antiviral therapy, and epidemiologic surveillance.
VA SeqFORCE laboratories use a variety of sequencing platforms, although a federated system was developed that electronically linked all laboratories using a software system (PraediGene, Bitscopic) for sample management, COVID-19 variant analytics, and automated result reporting of clade and lineage into the Veterans Health Information Systems and Technology Architecture (VistA) Computerized Patient Record System. In addition, generated nucleic acid sequence alignment through FASTA consensus sequence files have been archived for secondary research analyses. By archiving the consensus sequences, retrospective studies within the VA have the added benefit of being able to clinically annotate investigations into COVID-19 variant patterns. As of August 2023, 43,003 samples containing COVID-19 have been sequenced, and FASTA file and metadata upload are ongoing to the Global Initiative on Sharing Avian Influenza Data, which houses > 15 million COVID-19 files from global submissions.
VA SeqFORCE’s clinical sequencing efforts have created opportunities for multicenter collaboration in variant surveillance. In work from December 2021, investigators from the James J. Peters VA Medical Center in Bronx, New York, collaborated with the VHA Pathology and Laboratory Medicine Services and Public Health national program offices in Washington, DC, to develop an RT-PCR assay to rapidly differentiate Omicron from Delta variants.4 Samples from VA hospitals across the nation were used in this study.
Lessons from VA SeqFORCE have also been cited as inspiration to address COVID-19 clinical problems, including outbreak investigations in hospital settings and beyond. Researchers at the Iowa City VA Health Care System, for example, proposed a novel probabilistic quantitative method for determining genetic-relatedness among COVID-19 viral strains in an outbreak setting.5 They extended the scope of work to develop COVID-19 outbreak screening tools combining publicly available algorithms with targeted sequencing data to identify outbreaks as they arise.6 We expect VA SeqFORCE, in conjunction with its complement VA SeqCURE, will continue to further pandemic surveillance and response.
VA Seq CURE
As the research-focused complement to VA SeqFORCE, VA SeqCURE is dedicated to a broader study of the COVID-19 genome through sequencing. Established January 2021, the VA SeqCURE network consists of 6 research laboratories in Boise, Idaho; Bronx, New York; Cleveland, Ohio; Durham, North Carolina; Iowa City, Iowa; and Temple, Texas.
Samples are collected as a subset of the broader VA Science and Health Initiative to Combat Infectious and Emerging Life-Threatening Diseases (VA SHIELD) biorepository sweep protocol for discarded blood and nasal swab specimens of VHA patients hospitalized with COVID-19, as described by Epstein and colleagues.7-9 While VA SeqFORCE sequences samples positive for COVID-19 by RT-PCR with a Ct value of ≤ 30 for diagnostic purposes, VA SeqCURE laboratories sequence more broadly for nondiagnostic purposes, including samples with a Ct value > 30. The 6 VA SeqCURE laboratories generate sequencing data using various platforms, amplification kits, and formats. To ensure maximum quality and metadata on the sequences generated across the different laboratories, a sequence intake pipeline has been developed, adapting the ViralRecon bioinformatics platform.10 This harmonized analysis pipeline accommodates different file formats and performs quality control, alignment, variant calling, lineage assignment, clade assignment, and annotation. As of August 2023, VA SeqCURE has identified viral sequences from 24,107 unique specimens. Annotated COVID-19 sequences with the appropriate metadata will be available to VA researchers through VA SHIELD.
Research projects include descriptive epidemiology of COVID-19 variants in individuals who receive VHA care, COVID-19 vaccine and therapy effectiveness, and the unique distribution of variants and vaccine effectiveness in rural settings.3 True to its core mission, members of the VA SeqCURE consortium have contributed to the COVID-19 viral sequencing literature over the past 2 years. Researchers also are accessing VA SeqCURE to study COVID-19 persistence and rebound among individuals with mild disease taking nirmatrelvir/ritonavir compared with other COVID-19 therapeutics and untreated controls. Finally, COVID-19 samples and their sequences are stored in the VA SHIELD biorepository, which leverages these samples and data to advance scientific understanding of COVID-19 and future emerging infectious diseases.7-9
Important work from investigators at the Central Texas Veterans Health Care System confronted the issue of whole genome sequencing data from COVID-19 samples with low viral loads, a common issue with COVID-19 sequencing. They found that yields of 2 sequencing protocols, which generated high-sequence coverage, were enhanced further by combining the results of both methods.11 This project, which has potentially broad applications for sequencing in research and clinical settings, is an example of VA SeqCURE’s efforts to address the COVID-19 pandemic. The VA SeqCURE program has substantial potential as a large viral sequencing repository with broad geographic and demographic representation, such that future large-scale sequencing analyses may be generated from preexisting nested cohorts within the repository.
NEXT STEPS
Promising new directions of clinical and laboratory-based research are planned for VA SeqFORCE and VA SeqCURE. While the impact of COVID-19 and other viruses with epidemic potential is perhaps most feared in urban settings, evidence suggests that the distribution of COVID-19 in rural settings is unique and associated with worse outcomes.12,13 Given the wide catchment areas of VA hospitals that encompass both rural and urban settings, the VA’s ongoing COVID-19 sequencing programs and repositories are uniquely positioned to understand viral dynamics in areas of differing population density.
While rates of infection, hospitalization, and death resulting from COVID-19 have substantially dropped, the long-term impact of the pandemic is just beginning to be recognized in conditions such as long COVID or postacute COVID-19 syndrome. Long COVID has already proven to be biologically multifaceted, difficult to diagnose, and unpredictable in identifying the most at-risk patients.14-16 Much remains to be determined in our understanding of long COVID, including a unified definition that can effectively be used in clinical settings to diagnose and treat patients. However, research indicates that comorbidities common in veterans, such as diabetes and cardiovascular disease, are associated with worse long-term outcomes.17,18 Collaborations between VA scientists, clinicians, and national cooperative programs (such as a network of VHA long COVID clinics) create an unmatched opportunity for VA SeqFORCE and VA SeqCURE programs to provide insight into a disease likely to become a chronic disease outcome of the pandemic.
With VA SeqFORCE and VA SeqCURE programs, the VA now has infrastructure ready to respond to new infectious diseases. During the mpox outbreak of 2022, the VA Public Health Reference Laboratory received > 80% of all VA mpox samples for orthopox screening and mpox confirmatory testing. A subset of these samples underwent whole genome sequencing with the identification of 10 unique lineages across VA, and > 200 positive and 400 negative samples have been aliquoted and submitted to VA SHIELD for research. Furthermore, the VA SeqFORCE and VA SeqCURE sequencing processes might be adapted to identify outbreaks of multidrug-resistant organisms among VA patients trialed at other institutions.19 We are hopeful that VA SeqFORCE and VA SeqCURE will become invaluable components of health care delivery and infection prevention at the hospital level and beyond.
Finally, the robust data infrastructure and associated repositories of VA SeqFORCE and VA SeqCURE may be leveraged to study noninfectious diseases. Research groups are starting to apply these programs to cancer sequencing. We anticipate that these efforts may have a substantial impact on our understanding of cancer epidemiology and region-specific risk factors for malignancy, given the size and breadth of VA SeqFORCE and VA SeqCURE. Common oncogenic mutations identified through these programs could be targets for precision oncology therapeutics. Similarly, we envision applications of the VA SeqFORCE and VA SeqCURE data infrastructures and repositories toward other precision medicine fields, including pharmacogenomics and nutrition, to tailor interventions to meet the specific individual needs of veterans.
CONCLUSIONS
The productivity of VA SeqFORCE and VA SeqCURE programs over the past 2 years continues to increase in response to the COVID-19 pandemic. We anticipate that they will be vital components in our nation’s responses to infectious threats and beyond.
1. Iuliano AD, Brunkard JM, Boehmer TK, et al. Trends in disease severity and health care utilization during the early Omicron variant period compared with previous SARS-CoV-2 high transmission periods - United States, December 2020-January 2022. MMWR Morb Mortal Wkly Rep. 2022;71(4):146-152. Published 2022 Jan 28. doi:10.15585/mmwr.mm7104e4
2. Nyberg T, Ferguson NM, Nash SG, et al. Comparative analysis of the risks of hospitalisation and death associated with SARS-CoV-2 omicron (B.1.1.529) and delta (B.1.617.2) variants in England: a cohort study. Lancet. 2022;399(10332):1303-1312. doi:10.1016/S0140-6736(22)00462-7
3. Veterans Health Administration. Coronavirus Disease 2019 (COVID-19) response report - annex C. December 5, 2022. Accessed August 28, 2023. https://www.va.gov/HEALTH/docs/VHA-COVID-19-Response-2022-Annex-C.pdf 4. Barasch NJ, Iqbal J, Coombs M, et al. Utilization of a SARS-CoV-2 variant assay for the rapid differentiation of Omicron and Delta. medRxiv. Preprint posted online December 27, 2021. doi:10.1101/2021.12.22.21268195
5. Bilal MY. Similarity Index-probabilistic confidence estimation of SARS-CoV-2 strain relatedness in localized outbreaks. Epidemiologia (Basel). 2022;3(2):238-249. doi:10.3390/epidemiologia3020019
6. Bilal MY, Klutts JS. Molecular Epidemiological investigations of localized SARS-CoV-2 outbreaks-utility of public algorithms. Epidemiologia (Basel). 2022;3(3):402-411. doi:10.3390/epidemiologia3030031
7. Veterans Health Administration, Office of Research & Development. VA Science and Health Initiative to Combat Infectious and Emerging Life-Threatening Diseases (VA SHIELD). Updated November 23, 2022. Accessed August 28, 2023. https://www.research.va.gov/programs/shield/about.cfm
8. Harley JB, Pyarajan S, Partan ES, et al. The US Department of Veterans Affairs Science and Health Initiative to Combat Infectious and Emerging Life-Threatening Diseases (VA SHIELD): a biorepository addressing national health threats. Open Forum Infect Dis. 2022;9(12):ofac641. doi:10.1093/ofid/ofac641
9. Epstein L, Shive C, Garcia AP, et al. VA SHIELD: a biorepository for our veterans and the nation. Fed Pract. 2023;40(suppl 5):S48-S51. doi:10.12788/fp.0424
10. Patel H, Varona S, Monzón S, et al. Version 2.5. nf-core/viralrecon: nf-core/viralrecon v2.5 - Manganese Monkey (2.5). Zenodo. July 13, 2022. doi:10.5281/zenodo.6827984
11. Choi H, Hwang M, Navarathna DH, Xu J, Lukey J, Jinadatha C. Performance of COVIDSeq and swift normalase amplicon SARS-CoV-2 panels for SARS-CoV-2 genome sequencing: practical guide and combining FASTQ strategy. J Clin Microbiol. 2022;60(4):e0002522. doi:10.1128/jcm.00025-22
12. Cuadros DF, Branscum AJ, Mukandavire Z, Miller FD, MacKinnon N. Dynamics of the COVID-19 epidemic in urban and rural areas in the United States. Ann Epidemiol. 2021;59:16-20. doi:10.1016/j.annepidem.2021.04.007
13. Anzalone AJ, Horswell R, Hendricks BM, et al. Higher hospitalization and mortality rates among SARS-CoV-2-infected persons in rural America. J Rural Health. 2023;39(1):39-54. doi:10.1111/jrh.12689
14. Su Y, Yuan D, Chen DG, et al. Multiple early factors anticipate post-acute COVID-19 sequelae. Cell. 2022;185(5):881-895.e20. doi:10.1016/j.cell.2022.01.014
15. Pfaff ER, Girvin AT, Bennett TD, et al. Identifying who has long COVID in the USA: a machine learning approach using N3C data. Lancet Digit Health. 2022;4(7):e532-e541. doi:10.1016/S2589-7500(22)00048-6
16. Subramanian A, Nirantharakumar K, Hughes S, et al. Symptoms and risk factors for long COVID in non-hospitalized adults. Nat Med. 2022;28(8):1706-1714. doi:10.1038/s41591-022-01909-w
17. Munblit D, O’Hara ME, Akrami A, Perego E, Olliaro P, Needham DM. Long COVID: aiming for a consensus. Lancet Respir Med. 2022;10(7):632-634. doi:10.1016/S2213-2600(22)00135-7
18. Thaweethai T, Jolley SE, Karlson EW, et al. Development of a definition of postacute sequelae of SARS-CoV-2 infection. JAMA. 2023;329(22):1934-1946. doi:10.1001/jama.2023.8823
19. Sundermann AJ, Chen J, Kumar P, et al. Whole-genome sequencing surveillance and machine learning of the electronic health record for enhanced healthcare outbreak detection. Clin Infect Dis. 2022;75(3):476-482. doi:10.1093/cid/ciab946
The COVID-19 virus and its associated pandemic have highlighted the urgent need for a national infrastructure to rapidly identify and respond to emerging pathogens. The importance of understanding viral population dynamics through genetic sequencing has become apparent over time, particularly as the vaccine responses, clinical implications, and therapeutic effectiveness of treatments have varied substantially with COVID-19 variants.1,2
As the largest integrated health care system in the US, the US Department of Veterans Affairs (VA) is uniquely situated to help with pandemic detection and response. This article highlights 2 VA programs dedicated to COVID-19 sequencing at the forefront of pandemic response and research: VA Sequencing for Research Clinical and Epidemiology (SeqFORCE) and VA Sequencing Collaborations United for Research and Epidemiology (SeqCURE) (Table).
VA Seq FORCE
VA SeqFORCE was established March 2021 to facilitate clinical surveillance of COVID-19 variants in the US veteran population and in VA employees. VA SeqFORCE consists of 9 Clinical Laboratory Improvement Amendment (CLIA)–certified laboratories in VA medical centers, including the VA Public Health Reference Laboratory in Palo Alto, California, and 8 Veterans Health Administration (VHA) clinical laboratories (Los Angeles, California; Boise, Idaho; Iowa City, Iowa; Bronx, New York; West Haven, Connecticut; Indianapolis, Indiana; Denver, Colorado; and Orlando, Florida).3 Specimen standards (eg, real-time polymerase chain reaction [RT-PCR] cycle threshold [Ct] ≤ 30, minimum volume, etc) and clinical criteria (eg, COVID-19–related deaths, COVID-19 vaccine escape, etc) for submitting samples to VA SeqFORCE laboratories were established, and logistics for sample sequencing was centralized, including providing centralized instructions for sample preparation and to which VA SeqFORCE laboratory samples should be sent.
These laboratories sequenced samples from patients and employees with COVID-19 to understand patterns of variant evolution, vaccine, antiviral and monoclonal antibody response, health care–associated outbreaks, and COVID-19 transmission. As clinically relevant findings, such as monoclonal antibody treatment failure, emerged with novel viral variants, VA SeqFORCE was well positioned to rapidly detect the emergent variants and inform better clinical care of patients with COVID-19. Other clinical indications identified for sequencing within VA SeqFORCE included outbreak investigation, re-infection with COVID-19 > 90 days but < 6 months after a prior infection, extended hospitalization of > 21 days, death due to COVID-19, infection with a history of recent nondomestic travel, rebound of symptoms after improvement on oral antiviral therapy, and epidemiologic surveillance.
VA SeqFORCE laboratories use a variety of sequencing platforms, although a federated system was developed that electronically linked all laboratories using a software system (PraediGene, Bitscopic) for sample management, COVID-19 variant analytics, and automated result reporting of clade and lineage into the Veterans Health Information Systems and Technology Architecture (VistA) Computerized Patient Record System. In addition, generated nucleic acid sequence alignment through FASTA consensus sequence files have been archived for secondary research analyses. By archiving the consensus sequences, retrospective studies within the VA have the added benefit of being able to clinically annotate investigations into COVID-19 variant patterns. As of August 2023, 43,003 samples containing COVID-19 have been sequenced, and FASTA file and metadata upload are ongoing to the Global Initiative on Sharing Avian Influenza Data, which houses > 15 million COVID-19 files from global submissions.
VA SeqFORCE’s clinical sequencing efforts have created opportunities for multicenter collaboration in variant surveillance. In work from December 2021, investigators from the James J. Peters VA Medical Center in Bronx, New York, collaborated with the VHA Pathology and Laboratory Medicine Services and Public Health national program offices in Washington, DC, to develop an RT-PCR assay to rapidly differentiate Omicron from Delta variants.4 Samples from VA hospitals across the nation were used in this study.
Lessons from VA SeqFORCE have also been cited as inspiration to address COVID-19 clinical problems, including outbreak investigations in hospital settings and beyond. Researchers at the Iowa City VA Health Care System, for example, proposed a novel probabilistic quantitative method for determining genetic-relatedness among COVID-19 viral strains in an outbreak setting.5 They extended the scope of work to develop COVID-19 outbreak screening tools combining publicly available algorithms with targeted sequencing data to identify outbreaks as they arise.6 We expect VA SeqFORCE, in conjunction with its complement VA SeqCURE, will continue to further pandemic surveillance and response.
VA Seq CURE
As the research-focused complement to VA SeqFORCE, VA SeqCURE is dedicated to a broader study of the COVID-19 genome through sequencing. Established January 2021, the VA SeqCURE network consists of 6 research laboratories in Boise, Idaho; Bronx, New York; Cleveland, Ohio; Durham, North Carolina; Iowa City, Iowa; and Temple, Texas.
Samples are collected as a subset of the broader VA Science and Health Initiative to Combat Infectious and Emerging Life-Threatening Diseases (VA SHIELD) biorepository sweep protocol for discarded blood and nasal swab specimens of VHA patients hospitalized with COVID-19, as described by Epstein and colleagues.7-9 While VA SeqFORCE sequences samples positive for COVID-19 by RT-PCR with a Ct value of ≤ 30 for diagnostic purposes, VA SeqCURE laboratories sequence more broadly for nondiagnostic purposes, including samples with a Ct value > 30. The 6 VA SeqCURE laboratories generate sequencing data using various platforms, amplification kits, and formats. To ensure maximum quality and metadata on the sequences generated across the different laboratories, a sequence intake pipeline has been developed, adapting the ViralRecon bioinformatics platform.10 This harmonized analysis pipeline accommodates different file formats and performs quality control, alignment, variant calling, lineage assignment, clade assignment, and annotation. As of August 2023, VA SeqCURE has identified viral sequences from 24,107 unique specimens. Annotated COVID-19 sequences with the appropriate metadata will be available to VA researchers through VA SHIELD.
Research projects include descriptive epidemiology of COVID-19 variants in individuals who receive VHA care, COVID-19 vaccine and therapy effectiveness, and the unique distribution of variants and vaccine effectiveness in rural settings.3 True to its core mission, members of the VA SeqCURE consortium have contributed to the COVID-19 viral sequencing literature over the past 2 years. Researchers also are accessing VA SeqCURE to study COVID-19 persistence and rebound among individuals with mild disease taking nirmatrelvir/ritonavir compared with other COVID-19 therapeutics and untreated controls. Finally, COVID-19 samples and their sequences are stored in the VA SHIELD biorepository, which leverages these samples and data to advance scientific understanding of COVID-19 and future emerging infectious diseases.7-9
Important work from investigators at the Central Texas Veterans Health Care System confronted the issue of whole genome sequencing data from COVID-19 samples with low viral loads, a common issue with COVID-19 sequencing. They found that yields of 2 sequencing protocols, which generated high-sequence coverage, were enhanced further by combining the results of both methods.11 This project, which has potentially broad applications for sequencing in research and clinical settings, is an example of VA SeqCURE’s efforts to address the COVID-19 pandemic. The VA SeqCURE program has substantial potential as a large viral sequencing repository with broad geographic and demographic representation, such that future large-scale sequencing analyses may be generated from preexisting nested cohorts within the repository.
NEXT STEPS
Promising new directions of clinical and laboratory-based research are planned for VA SeqFORCE and VA SeqCURE. While the impact of COVID-19 and other viruses with epidemic potential is perhaps most feared in urban settings, evidence suggests that the distribution of COVID-19 in rural settings is unique and associated with worse outcomes.12,13 Given the wide catchment areas of VA hospitals that encompass both rural and urban settings, the VA’s ongoing COVID-19 sequencing programs and repositories are uniquely positioned to understand viral dynamics in areas of differing population density.
While rates of infection, hospitalization, and death resulting from COVID-19 have substantially dropped, the long-term impact of the pandemic is just beginning to be recognized in conditions such as long COVID or postacute COVID-19 syndrome. Long COVID has already proven to be biologically multifaceted, difficult to diagnose, and unpredictable in identifying the most at-risk patients.14-16 Much remains to be determined in our understanding of long COVID, including a unified definition that can effectively be used in clinical settings to diagnose and treat patients. However, research indicates that comorbidities common in veterans, such as diabetes and cardiovascular disease, are associated with worse long-term outcomes.17,18 Collaborations between VA scientists, clinicians, and national cooperative programs (such as a network of VHA long COVID clinics) create an unmatched opportunity for VA SeqFORCE and VA SeqCURE programs to provide insight into a disease likely to become a chronic disease outcome of the pandemic.
With VA SeqFORCE and VA SeqCURE programs, the VA now has infrastructure ready to respond to new infectious diseases. During the mpox outbreak of 2022, the VA Public Health Reference Laboratory received > 80% of all VA mpox samples for orthopox screening and mpox confirmatory testing. A subset of these samples underwent whole genome sequencing with the identification of 10 unique lineages across VA, and > 200 positive and 400 negative samples have been aliquoted and submitted to VA SHIELD for research. Furthermore, the VA SeqFORCE and VA SeqCURE sequencing processes might be adapted to identify outbreaks of multidrug-resistant organisms among VA patients trialed at other institutions.19 We are hopeful that VA SeqFORCE and VA SeqCURE will become invaluable components of health care delivery and infection prevention at the hospital level and beyond.
Finally, the robust data infrastructure and associated repositories of VA SeqFORCE and VA SeqCURE may be leveraged to study noninfectious diseases. Research groups are starting to apply these programs to cancer sequencing. We anticipate that these efforts may have a substantial impact on our understanding of cancer epidemiology and region-specific risk factors for malignancy, given the size and breadth of VA SeqFORCE and VA SeqCURE. Common oncogenic mutations identified through these programs could be targets for precision oncology therapeutics. Similarly, we envision applications of the VA SeqFORCE and VA SeqCURE data infrastructures and repositories toward other precision medicine fields, including pharmacogenomics and nutrition, to tailor interventions to meet the specific individual needs of veterans.
CONCLUSIONS
The productivity of VA SeqFORCE and VA SeqCURE programs over the past 2 years continues to increase in response to the COVID-19 pandemic. We anticipate that they will be vital components in our nation’s responses to infectious threats and beyond.
The COVID-19 virus and its associated pandemic have highlighted the urgent need for a national infrastructure to rapidly identify and respond to emerging pathogens. The importance of understanding viral population dynamics through genetic sequencing has become apparent over time, particularly as the vaccine responses, clinical implications, and therapeutic effectiveness of treatments have varied substantially with COVID-19 variants.1,2
As the largest integrated health care system in the US, the US Department of Veterans Affairs (VA) is uniquely situated to help with pandemic detection and response. This article highlights 2 VA programs dedicated to COVID-19 sequencing at the forefront of pandemic response and research: VA Sequencing for Research Clinical and Epidemiology (SeqFORCE) and VA Sequencing Collaborations United for Research and Epidemiology (SeqCURE) (Table).
VA Seq FORCE
VA SeqFORCE was established March 2021 to facilitate clinical surveillance of COVID-19 variants in the US veteran population and in VA employees. VA SeqFORCE consists of 9 Clinical Laboratory Improvement Amendment (CLIA)–certified laboratories in VA medical centers, including the VA Public Health Reference Laboratory in Palo Alto, California, and 8 Veterans Health Administration (VHA) clinical laboratories (Los Angeles, California; Boise, Idaho; Iowa City, Iowa; Bronx, New York; West Haven, Connecticut; Indianapolis, Indiana; Denver, Colorado; and Orlando, Florida).3 Specimen standards (eg, real-time polymerase chain reaction [RT-PCR] cycle threshold [Ct] ≤ 30, minimum volume, etc) and clinical criteria (eg, COVID-19–related deaths, COVID-19 vaccine escape, etc) for submitting samples to VA SeqFORCE laboratories were established, and logistics for sample sequencing was centralized, including providing centralized instructions for sample preparation and to which VA SeqFORCE laboratory samples should be sent.
These laboratories sequenced samples from patients and employees with COVID-19 to understand patterns of variant evolution, vaccine, antiviral and monoclonal antibody response, health care–associated outbreaks, and COVID-19 transmission. As clinically relevant findings, such as monoclonal antibody treatment failure, emerged with novel viral variants, VA SeqFORCE was well positioned to rapidly detect the emergent variants and inform better clinical care of patients with COVID-19. Other clinical indications identified for sequencing within VA SeqFORCE included outbreak investigation, re-infection with COVID-19 > 90 days but < 6 months after a prior infection, extended hospitalization of > 21 days, death due to COVID-19, infection with a history of recent nondomestic travel, rebound of symptoms after improvement on oral antiviral therapy, and epidemiologic surveillance.
VA SeqFORCE laboratories use a variety of sequencing platforms, although a federated system was developed that electronically linked all laboratories using a software system (PraediGene, Bitscopic) for sample management, COVID-19 variant analytics, and automated result reporting of clade and lineage into the Veterans Health Information Systems and Technology Architecture (VistA) Computerized Patient Record System. In addition, generated nucleic acid sequence alignment through FASTA consensus sequence files have been archived for secondary research analyses. By archiving the consensus sequences, retrospective studies within the VA have the added benefit of being able to clinically annotate investigations into COVID-19 variant patterns. As of August 2023, 43,003 samples containing COVID-19 have been sequenced, and FASTA file and metadata upload are ongoing to the Global Initiative on Sharing Avian Influenza Data, which houses > 15 million COVID-19 files from global submissions.
VA SeqFORCE’s clinical sequencing efforts have created opportunities for multicenter collaboration in variant surveillance. In work from December 2021, investigators from the James J. Peters VA Medical Center in Bronx, New York, collaborated with the VHA Pathology and Laboratory Medicine Services and Public Health national program offices in Washington, DC, to develop an RT-PCR assay to rapidly differentiate Omicron from Delta variants.4 Samples from VA hospitals across the nation were used in this study.
Lessons from VA SeqFORCE have also been cited as inspiration to address COVID-19 clinical problems, including outbreak investigations in hospital settings and beyond. Researchers at the Iowa City VA Health Care System, for example, proposed a novel probabilistic quantitative method for determining genetic-relatedness among COVID-19 viral strains in an outbreak setting.5 They extended the scope of work to develop COVID-19 outbreak screening tools combining publicly available algorithms with targeted sequencing data to identify outbreaks as they arise.6 We expect VA SeqFORCE, in conjunction with its complement VA SeqCURE, will continue to further pandemic surveillance and response.
VA Seq CURE
As the research-focused complement to VA SeqFORCE, VA SeqCURE is dedicated to a broader study of the COVID-19 genome through sequencing. Established January 2021, the VA SeqCURE network consists of 6 research laboratories in Boise, Idaho; Bronx, New York; Cleveland, Ohio; Durham, North Carolina; Iowa City, Iowa; and Temple, Texas.
Samples are collected as a subset of the broader VA Science and Health Initiative to Combat Infectious and Emerging Life-Threatening Diseases (VA SHIELD) biorepository sweep protocol for discarded blood and nasal swab specimens of VHA patients hospitalized with COVID-19, as described by Epstein and colleagues.7-9 While VA SeqFORCE sequences samples positive for COVID-19 by RT-PCR with a Ct value of ≤ 30 for diagnostic purposes, VA SeqCURE laboratories sequence more broadly for nondiagnostic purposes, including samples with a Ct value > 30. The 6 VA SeqCURE laboratories generate sequencing data using various platforms, amplification kits, and formats. To ensure maximum quality and metadata on the sequences generated across the different laboratories, a sequence intake pipeline has been developed, adapting the ViralRecon bioinformatics platform.10 This harmonized analysis pipeline accommodates different file formats and performs quality control, alignment, variant calling, lineage assignment, clade assignment, and annotation. As of August 2023, VA SeqCURE has identified viral sequences from 24,107 unique specimens. Annotated COVID-19 sequences with the appropriate metadata will be available to VA researchers through VA SHIELD.
Research projects include descriptive epidemiology of COVID-19 variants in individuals who receive VHA care, COVID-19 vaccine and therapy effectiveness, and the unique distribution of variants and vaccine effectiveness in rural settings.3 True to its core mission, members of the VA SeqCURE consortium have contributed to the COVID-19 viral sequencing literature over the past 2 years. Researchers also are accessing VA SeqCURE to study COVID-19 persistence and rebound among individuals with mild disease taking nirmatrelvir/ritonavir compared with other COVID-19 therapeutics and untreated controls. Finally, COVID-19 samples and their sequences are stored in the VA SHIELD biorepository, which leverages these samples and data to advance scientific understanding of COVID-19 and future emerging infectious diseases.7-9
Important work from investigators at the Central Texas Veterans Health Care System confronted the issue of whole genome sequencing data from COVID-19 samples with low viral loads, a common issue with COVID-19 sequencing. They found that yields of 2 sequencing protocols, which generated high-sequence coverage, were enhanced further by combining the results of both methods.11 This project, which has potentially broad applications for sequencing in research and clinical settings, is an example of VA SeqCURE’s efforts to address the COVID-19 pandemic. The VA SeqCURE program has substantial potential as a large viral sequencing repository with broad geographic and demographic representation, such that future large-scale sequencing analyses may be generated from preexisting nested cohorts within the repository.
NEXT STEPS
Promising new directions of clinical and laboratory-based research are planned for VA SeqFORCE and VA SeqCURE. While the impact of COVID-19 and other viruses with epidemic potential is perhaps most feared in urban settings, evidence suggests that the distribution of COVID-19 in rural settings is unique and associated with worse outcomes.12,13 Given the wide catchment areas of VA hospitals that encompass both rural and urban settings, the VA’s ongoing COVID-19 sequencing programs and repositories are uniquely positioned to understand viral dynamics in areas of differing population density.
While rates of infection, hospitalization, and death resulting from COVID-19 have substantially dropped, the long-term impact of the pandemic is just beginning to be recognized in conditions such as long COVID or postacute COVID-19 syndrome. Long COVID has already proven to be biologically multifaceted, difficult to diagnose, and unpredictable in identifying the most at-risk patients.14-16 Much remains to be determined in our understanding of long COVID, including a unified definition that can effectively be used in clinical settings to diagnose and treat patients. However, research indicates that comorbidities common in veterans, such as diabetes and cardiovascular disease, are associated with worse long-term outcomes.17,18 Collaborations between VA scientists, clinicians, and national cooperative programs (such as a network of VHA long COVID clinics) create an unmatched opportunity for VA SeqFORCE and VA SeqCURE programs to provide insight into a disease likely to become a chronic disease outcome of the pandemic.
With VA SeqFORCE and VA SeqCURE programs, the VA now has infrastructure ready to respond to new infectious diseases. During the mpox outbreak of 2022, the VA Public Health Reference Laboratory received > 80% of all VA mpox samples for orthopox screening and mpox confirmatory testing. A subset of these samples underwent whole genome sequencing with the identification of 10 unique lineages across VA, and > 200 positive and 400 negative samples have been aliquoted and submitted to VA SHIELD for research. Furthermore, the VA SeqFORCE and VA SeqCURE sequencing processes might be adapted to identify outbreaks of multidrug-resistant organisms among VA patients trialed at other institutions.19 We are hopeful that VA SeqFORCE and VA SeqCURE will become invaluable components of health care delivery and infection prevention at the hospital level and beyond.
Finally, the robust data infrastructure and associated repositories of VA SeqFORCE and VA SeqCURE may be leveraged to study noninfectious diseases. Research groups are starting to apply these programs to cancer sequencing. We anticipate that these efforts may have a substantial impact on our understanding of cancer epidemiology and region-specific risk factors for malignancy, given the size and breadth of VA SeqFORCE and VA SeqCURE. Common oncogenic mutations identified through these programs could be targets for precision oncology therapeutics. Similarly, we envision applications of the VA SeqFORCE and VA SeqCURE data infrastructures and repositories toward other precision medicine fields, including pharmacogenomics and nutrition, to tailor interventions to meet the specific individual needs of veterans.
CONCLUSIONS
The productivity of VA SeqFORCE and VA SeqCURE programs over the past 2 years continues to increase in response to the COVID-19 pandemic. We anticipate that they will be vital components in our nation’s responses to infectious threats and beyond.
1. Iuliano AD, Brunkard JM, Boehmer TK, et al. Trends in disease severity and health care utilization during the early Omicron variant period compared with previous SARS-CoV-2 high transmission periods - United States, December 2020-January 2022. MMWR Morb Mortal Wkly Rep. 2022;71(4):146-152. Published 2022 Jan 28. doi:10.15585/mmwr.mm7104e4
2. Nyberg T, Ferguson NM, Nash SG, et al. Comparative analysis of the risks of hospitalisation and death associated with SARS-CoV-2 omicron (B.1.1.529) and delta (B.1.617.2) variants in England: a cohort study. Lancet. 2022;399(10332):1303-1312. doi:10.1016/S0140-6736(22)00462-7
3. Veterans Health Administration. Coronavirus Disease 2019 (COVID-19) response report - annex C. December 5, 2022. Accessed August 28, 2023. https://www.va.gov/HEALTH/docs/VHA-COVID-19-Response-2022-Annex-C.pdf 4. Barasch NJ, Iqbal J, Coombs M, et al. Utilization of a SARS-CoV-2 variant assay for the rapid differentiation of Omicron and Delta. medRxiv. Preprint posted online December 27, 2021. doi:10.1101/2021.12.22.21268195
5. Bilal MY. Similarity Index-probabilistic confidence estimation of SARS-CoV-2 strain relatedness in localized outbreaks. Epidemiologia (Basel). 2022;3(2):238-249. doi:10.3390/epidemiologia3020019
6. Bilal MY, Klutts JS. Molecular Epidemiological investigations of localized SARS-CoV-2 outbreaks-utility of public algorithms. Epidemiologia (Basel). 2022;3(3):402-411. doi:10.3390/epidemiologia3030031
7. Veterans Health Administration, Office of Research & Development. VA Science and Health Initiative to Combat Infectious and Emerging Life-Threatening Diseases (VA SHIELD). Updated November 23, 2022. Accessed August 28, 2023. https://www.research.va.gov/programs/shield/about.cfm
8. Harley JB, Pyarajan S, Partan ES, et al. The US Department of Veterans Affairs Science and Health Initiative to Combat Infectious and Emerging Life-Threatening Diseases (VA SHIELD): a biorepository addressing national health threats. Open Forum Infect Dis. 2022;9(12):ofac641. doi:10.1093/ofid/ofac641
9. Epstein L, Shive C, Garcia AP, et al. VA SHIELD: a biorepository for our veterans and the nation. Fed Pract. 2023;40(suppl 5):S48-S51. doi:10.12788/fp.0424
10. Patel H, Varona S, Monzón S, et al. Version 2.5. nf-core/viralrecon: nf-core/viralrecon v2.5 - Manganese Monkey (2.5). Zenodo. July 13, 2022. doi:10.5281/zenodo.6827984
11. Choi H, Hwang M, Navarathna DH, Xu J, Lukey J, Jinadatha C. Performance of COVIDSeq and swift normalase amplicon SARS-CoV-2 panels for SARS-CoV-2 genome sequencing: practical guide and combining FASTQ strategy. J Clin Microbiol. 2022;60(4):e0002522. doi:10.1128/jcm.00025-22
12. Cuadros DF, Branscum AJ, Mukandavire Z, Miller FD, MacKinnon N. Dynamics of the COVID-19 epidemic in urban and rural areas in the United States. Ann Epidemiol. 2021;59:16-20. doi:10.1016/j.annepidem.2021.04.007
13. Anzalone AJ, Horswell R, Hendricks BM, et al. Higher hospitalization and mortality rates among SARS-CoV-2-infected persons in rural America. J Rural Health. 2023;39(1):39-54. doi:10.1111/jrh.12689
14. Su Y, Yuan D, Chen DG, et al. Multiple early factors anticipate post-acute COVID-19 sequelae. Cell. 2022;185(5):881-895.e20. doi:10.1016/j.cell.2022.01.014
15. Pfaff ER, Girvin AT, Bennett TD, et al. Identifying who has long COVID in the USA: a machine learning approach using N3C data. Lancet Digit Health. 2022;4(7):e532-e541. doi:10.1016/S2589-7500(22)00048-6
16. Subramanian A, Nirantharakumar K, Hughes S, et al. Symptoms and risk factors for long COVID in non-hospitalized adults. Nat Med. 2022;28(8):1706-1714. doi:10.1038/s41591-022-01909-w
17. Munblit D, O’Hara ME, Akrami A, Perego E, Olliaro P, Needham DM. Long COVID: aiming for a consensus. Lancet Respir Med. 2022;10(7):632-634. doi:10.1016/S2213-2600(22)00135-7
18. Thaweethai T, Jolley SE, Karlson EW, et al. Development of a definition of postacute sequelae of SARS-CoV-2 infection. JAMA. 2023;329(22):1934-1946. doi:10.1001/jama.2023.8823
19. Sundermann AJ, Chen J, Kumar P, et al. Whole-genome sequencing surveillance and machine learning of the electronic health record for enhanced healthcare outbreak detection. Clin Infect Dis. 2022;75(3):476-482. doi:10.1093/cid/ciab946
1. Iuliano AD, Brunkard JM, Boehmer TK, et al. Trends in disease severity and health care utilization during the early Omicron variant period compared with previous SARS-CoV-2 high transmission periods - United States, December 2020-January 2022. MMWR Morb Mortal Wkly Rep. 2022;71(4):146-152. Published 2022 Jan 28. doi:10.15585/mmwr.mm7104e4
2. Nyberg T, Ferguson NM, Nash SG, et al. Comparative analysis of the risks of hospitalisation and death associated with SARS-CoV-2 omicron (B.1.1.529) and delta (B.1.617.2) variants in England: a cohort study. Lancet. 2022;399(10332):1303-1312. doi:10.1016/S0140-6736(22)00462-7
3. Veterans Health Administration. Coronavirus Disease 2019 (COVID-19) response report - annex C. December 5, 2022. Accessed August 28, 2023. https://www.va.gov/HEALTH/docs/VHA-COVID-19-Response-2022-Annex-C.pdf 4. Barasch NJ, Iqbal J, Coombs M, et al. Utilization of a SARS-CoV-2 variant assay for the rapid differentiation of Omicron and Delta. medRxiv. Preprint posted online December 27, 2021. doi:10.1101/2021.12.22.21268195
5. Bilal MY. Similarity Index-probabilistic confidence estimation of SARS-CoV-2 strain relatedness in localized outbreaks. Epidemiologia (Basel). 2022;3(2):238-249. doi:10.3390/epidemiologia3020019
6. Bilal MY, Klutts JS. Molecular Epidemiological investigations of localized SARS-CoV-2 outbreaks-utility of public algorithms. Epidemiologia (Basel). 2022;3(3):402-411. doi:10.3390/epidemiologia3030031
7. Veterans Health Administration, Office of Research & Development. VA Science and Health Initiative to Combat Infectious and Emerging Life-Threatening Diseases (VA SHIELD). Updated November 23, 2022. Accessed August 28, 2023. https://www.research.va.gov/programs/shield/about.cfm
8. Harley JB, Pyarajan S, Partan ES, et al. The US Department of Veterans Affairs Science and Health Initiative to Combat Infectious and Emerging Life-Threatening Diseases (VA SHIELD): a biorepository addressing national health threats. Open Forum Infect Dis. 2022;9(12):ofac641. doi:10.1093/ofid/ofac641
9. Epstein L, Shive C, Garcia AP, et al. VA SHIELD: a biorepository for our veterans and the nation. Fed Pract. 2023;40(suppl 5):S48-S51. doi:10.12788/fp.0424
10. Patel H, Varona S, Monzón S, et al. Version 2.5. nf-core/viralrecon: nf-core/viralrecon v2.5 - Manganese Monkey (2.5). Zenodo. July 13, 2022. doi:10.5281/zenodo.6827984
11. Choi H, Hwang M, Navarathna DH, Xu J, Lukey J, Jinadatha C. Performance of COVIDSeq and swift normalase amplicon SARS-CoV-2 panels for SARS-CoV-2 genome sequencing: practical guide and combining FASTQ strategy. J Clin Microbiol. 2022;60(4):e0002522. doi:10.1128/jcm.00025-22
12. Cuadros DF, Branscum AJ, Mukandavire Z, Miller FD, MacKinnon N. Dynamics of the COVID-19 epidemic in urban and rural areas in the United States. Ann Epidemiol. 2021;59:16-20. doi:10.1016/j.annepidem.2021.04.007
13. Anzalone AJ, Horswell R, Hendricks BM, et al. Higher hospitalization and mortality rates among SARS-CoV-2-infected persons in rural America. J Rural Health. 2023;39(1):39-54. doi:10.1111/jrh.12689
14. Su Y, Yuan D, Chen DG, et al. Multiple early factors anticipate post-acute COVID-19 sequelae. Cell. 2022;185(5):881-895.e20. doi:10.1016/j.cell.2022.01.014
15. Pfaff ER, Girvin AT, Bennett TD, et al. Identifying who has long COVID in the USA: a machine learning approach using N3C data. Lancet Digit Health. 2022;4(7):e532-e541. doi:10.1016/S2589-7500(22)00048-6
16. Subramanian A, Nirantharakumar K, Hughes S, et al. Symptoms and risk factors for long COVID in non-hospitalized adults. Nat Med. 2022;28(8):1706-1714. doi:10.1038/s41591-022-01909-w
17. Munblit D, O’Hara ME, Akrami A, Perego E, Olliaro P, Needham DM. Long COVID: aiming for a consensus. Lancet Respir Med. 2022;10(7):632-634. doi:10.1016/S2213-2600(22)00135-7
18. Thaweethai T, Jolley SE, Karlson EW, et al. Development of a definition of postacute sequelae of SARS-CoV-2 infection. JAMA. 2023;329(22):1934-1946. doi:10.1001/jama.2023.8823
19. Sundermann AJ, Chen J, Kumar P, et al. Whole-genome sequencing surveillance and machine learning of the electronic health record for enhanced healthcare outbreak detection. Clin Infect Dis. 2022;75(3):476-482. doi:10.1093/cid/ciab946
